Generalization to Bayesian Softmax Regression
Contents
Generalization to Bayesian Softmax Regression#
%pip install pymc pytensor
Requirement already satisfied: pymc in /usr/local/lib/python3.10/dist-packages (5.7.2)
Requirement already satisfied: pytensor in /usr/local/lib/python3.10/dist-packages (2.14.2)
Requirement already satisfied: arviz>=0.13.0 in /usr/local/lib/python3.10/dist-packages (from pymc) (0.15.1)
Requirement already satisfied: cachetools>=4.2.1 in /usr/local/lib/python3.10/dist-packages (from pymc) (5.3.2)
Requirement already satisfied: cloudpickle in /usr/local/lib/python3.10/dist-packages (from pymc) (2.2.1)
Requirement already satisfied: fastprogress>=0.2.0 in /usr/local/lib/python3.10/dist-packages (from pymc) (1.0.3)
Requirement already satisfied: numpy>=1.15.0 in /usr/local/lib/python3.10/dist-packages (from pymc) (1.25.2)
Requirement already satisfied: pandas>=0.24.0 in /usr/local/lib/python3.10/dist-packages (from pymc) (1.5.3)
Requirement already satisfied: scipy>=1.4.1 in /usr/local/lib/python3.10/dist-packages (from pymc) (1.11.4)
Requirement already satisfied: typing-extensions>=3.7.4 in /usr/local/lib/python3.10/dist-packages (from pymc) (4.9.0)
Requirement already satisfied: setuptools>=48.0.0 in /usr/local/lib/python3.10/dist-packages (from pytensor) (67.7.2)
Requirement already satisfied: filelock in /usr/local/lib/python3.10/dist-packages (from pytensor) (3.13.1)
Requirement already satisfied: etuples in /usr/local/lib/python3.10/dist-packages (from pytensor) (0.3.9)
Requirement already satisfied: logical-unification in /usr/local/lib/python3.10/dist-packages (from pytensor) (0.4.6)
Requirement already satisfied: miniKanren in /usr/local/lib/python3.10/dist-packages (from pytensor) (1.0.3)
Requirement already satisfied: cons in /usr/local/lib/python3.10/dist-packages (from pytensor) (0.4.6)
Requirement already satisfied: matplotlib>=3.2 in /usr/local/lib/python3.10/dist-packages (from arviz>=0.13.0->pymc) (3.7.1)
Requirement already satisfied: packaging in /usr/local/lib/python3.10/dist-packages (from arviz>=0.13.0->pymc) (23.2)
Requirement already satisfied: xarray>=0.21.0 in /usr/local/lib/python3.10/dist-packages (from arviz>=0.13.0->pymc) (2023.7.0)
Requirement already satisfied: h5netcdf>=1.0.2 in /usr/local/lib/python3.10/dist-packages (from arviz>=0.13.0->pymc) (1.3.0)
Requirement already satisfied: xarray-einstats>=0.3 in /usr/local/lib/python3.10/dist-packages (from arviz>=0.13.0->pymc) (0.7.0)
Requirement already satisfied: python-dateutil>=2.8.1 in /usr/local/lib/python3.10/dist-packages (from pandas>=0.24.0->pymc) (2.8.2)
Requirement already satisfied: pytz>=2020.1 in /usr/local/lib/python3.10/dist-packages (from pandas>=0.24.0->pymc) (2023.4)
Requirement already satisfied: toolz in /usr/local/lib/python3.10/dist-packages (from logical-unification->pytensor) (0.12.1)
Requirement already satisfied: multipledispatch in /usr/local/lib/python3.10/dist-packages (from logical-unification->pytensor) (1.0.0)
Requirement already satisfied: h5py in /usr/local/lib/python3.10/dist-packages (from h5netcdf>=1.0.2->arviz>=0.13.0->pymc) (3.9.0)
Requirement already satisfied: contourpy>=1.0.1 in /usr/local/lib/python3.10/dist-packages (from matplotlib>=3.2->arviz>=0.13.0->pymc) (1.2.0)
Requirement already satisfied: cycler>=0.10 in /usr/local/lib/python3.10/dist-packages (from matplotlib>=3.2->arviz>=0.13.0->pymc) (0.12.1)
Requirement already satisfied: fonttools>=4.22.0 in /usr/local/lib/python3.10/dist-packages (from matplotlib>=3.2->arviz>=0.13.0->pymc) (4.49.0)
Requirement already satisfied: kiwisolver>=1.0.1 in /usr/local/lib/python3.10/dist-packages (from matplotlib>=3.2->arviz>=0.13.0->pymc) (1.4.5)
Requirement already satisfied: pillow>=6.2.0 in /usr/local/lib/python3.10/dist-packages (from matplotlib>=3.2->arviz>=0.13.0->pymc) (9.4.0)
Requirement already satisfied: pyparsing>=2.3.1 in /usr/local/lib/python3.10/dist-packages (from matplotlib>=3.2->arviz>=0.13.0->pymc) (3.1.1)
Requirement already satisfied: six>=1.5 in /usr/local/lib/python3.10/dist-packages (from python-dateutil>=2.8.1->pandas>=0.24.0->pymc) (1.16.0)
import pymc as pm
import numpy as np
import pandas as pd
import scipy.stats as stats
from scipy.special import expit as logistic
import matplotlib.pyplot as plt
import arviz as az
import requests
import io
az.style.use('arviz-darkgrid')
target_url = 'https://raw.githubusercontent.com/cfteach/brds/main/datasets/iris.csv'
download = requests.get(target_url).content
iris = pd.read_csv(io.StringIO(download.decode('utf-8')))
iris.head()
| sepal_length | sepal_width | petal_length | petal_width | species | |
|---|---|---|---|---|---|
| 0 | 5.1 | 3.5 | 1.4 | 0.2 | setosa |
| 1 | 4.9 | 3.0 | 1.4 | 0.2 | setosa |
| 2 | 4.7 | 3.2 | 1.3 | 0.2 | setosa |
| 3 | 4.6 | 3.1 | 1.5 | 0.2 | setosa |
| 4 | 5.0 | 3.6 | 1.4 | 0.2 | setosa |
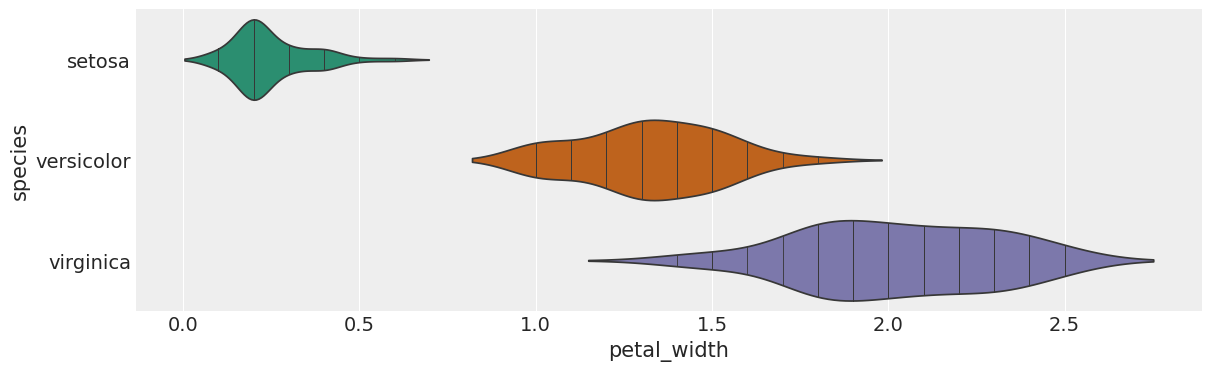
# @title species vs petal_width
from matplotlib import pyplot as plt
import seaborn as sns
figsize = (12, 1.2 * len(iris['species'].unique()))
plt.figure(figsize=figsize)
sns.violinplot(iris, x='petal_width', y='species', inner='stick', palette='Dark2')
sns.despine(top=True, right=True, bottom=True, left=True)
<ipython-input-6-7a5e472054ba>:7: FutureWarning:
Passing `palette` without assigning `hue` is deprecated and will be removed in v0.14.0. Assign the `y` variable to `hue` and set `legend=False` for the same effect.
sns.violinplot(iris, x='petal_width', y='species', inner='stick', palette='Dark2')
Generalization to multiple classes: Softmax Regression#
Notes
In order to generalize to mutliple classes, two modifications are needed:
(i) We use a softmax (see also Boltzmann distribution in physics), which is defined as:
\(softmax_{i}(\mu)= \frac{exp(\mu_{i})}{\sum_{k}exp(\mu_{k})}\)
(ii) We then replace the Bernoulli distribution with the categorical distribution. As the Bernoulli (single coin flip) is a special case of a Binomial (n coin flips), the categorical (single roll of a die) is a special case of the multinomial distribution (n rolls of a die).
iris.columns[:-1]
Index(['sepal_length', 'sepal_width', 'petal_length', 'petal_width'], dtype='object')
iris = sns.load_dataset('iris')
y_s = pd.Categorical(iris['species']).codes
x_n = iris.columns[:-1]
x_s = iris[x_n].values
x_s = (x_s - x_s.mean(axis=0)) / x_s.std(axis=0)
print(np.shape(x_s))
print(np.shape(y_s))
#print(x_n)
#print(iris)
(150, 4)
(150,)
with pm.Model() as model_s:
α = pm.Normal('α', mu=0, sigma=5, shape=3)
β = pm.Normal('β', mu=0, sigma=5, shape=(4,3))
μ = pm.Deterministic('μ', α + pm.math.dot(x_s, β))
θ = pm.math.softmax(μ, axis = 1)
yl = pm.Categorical('yl', p=θ, observed=y_s)
idata_s = pm.sample(2000, target_accept=.95)
Note that you need to call .eval() on Theano tensors to convert them to NumPy arrays before passing them to the softmax function.
data_pred = idata_s.posterior['μ'].mean(axis=0).mean(axis=0).values
print(np.shape(data_pred))
print(np.shape(y_s))
(150, 3)
(150,)
y_pred = [np.exp(point)/np.sum(np.exp(point), axis=0)
for point in data_pred]
res_t = np.sum(y_s == np.argmax(y_pred, axis=1)) / len(y_s)
print("accuracy is: {:1.3f}".format(res_t))
accuracy is: 0.980
az.summary(idata_s).head(15)
/usr/local/lib/python3.10/dist-packages/arviz/utils.py:184: NumbaDeprecationWarning: The 'nopython' keyword argument was not supplied to the 'numba.jit' decorator. The implicit default value for this argument is currently False, but it will be changed to True in Numba 0.59.0. See https://numba.readthedocs.io/en/stable/reference/deprecation.html#deprecation-of-object-mode-fall-back-behaviour-when-using-jit for details.
numba_fn = numba.jit(**self.kwargs)(self.function)
| mean | sd | hdi_3% | hdi_97% | mcse_mean | mcse_sd | ess_bulk | ess_tail | r_hat | |
|---|---|---|---|---|---|---|---|---|---|
| α[0] | -1.128 | 3.766 | -7.880 | 6.266 | 0.066 | 0.054 | 3236.0 | 2625.0 | 1.0 |
| α[1] | 5.855 | 3.194 | 0.224 | 12.107 | 0.060 | 0.043 | 2830.0 | 2885.0 | 1.0 |
| α[2] | -4.826 | 3.432 | -11.594 | 1.316 | 0.061 | 0.045 | 3133.0 | 2971.0 | 1.0 |
| β[0, 0] | -2.461 | 4.161 | -10.526 | 5.205 | 0.067 | 0.059 | 3794.0 | 3053.0 | 1.0 |
| β[0, 1] | 1.970 | 3.316 | -4.186 | 8.424 | 0.068 | 0.050 | 2369.0 | 2622.0 | 1.0 |
| β[0, 2] | 0.606 | 3.332 | -5.372 | 7.274 | 0.069 | 0.052 | 2312.0 | 2244.0 | 1.0 |
| β[1, 0] | 3.258 | 3.395 | -2.710 | 9.906 | 0.066 | 0.048 | 2678.0 | 2749.0 | 1.0 |
| β[1, 1] | -0.864 | 3.025 | -6.356 | 5.093 | 0.064 | 0.046 | 2235.0 | 2242.0 | 1.0 |
| β[1, 2] | -2.268 | 3.050 | -8.011 | 3.336 | 0.065 | 0.046 | 2227.0 | 2475.0 | 1.0 |
| β[2, 0] | -6.319 | 4.300 | -14.562 | 1.564 | 0.071 | 0.054 | 3708.0 | 2755.0 | 1.0 |
| β[2, 1] | -1.555 | 3.654 | -8.427 | 5.317 | 0.062 | 0.054 | 3526.0 | 3002.0 | 1.0 |
| β[2, 2] | 7.660 | 3.772 | 0.598 | 14.655 | 0.061 | 0.044 | 3796.0 | 3133.0 | 1.0 |
| β[3, 0] | -5.829 | 4.248 | -14.152 | 2.004 | 0.074 | 0.055 | 3292.0 | 3136.0 | 1.0 |
| β[3, 1] | -0.888 | 3.486 | -7.276 | 5.645 | 0.068 | 0.049 | 2625.0 | 3028.0 | 1.0 |
| β[3, 2] | 6.956 | 3.616 | 0.164 | 13.683 | 0.070 | 0.051 | 2697.0 | 2893.0 | 1.0 |
Notes
(i) 98% is the accuracy on our data; a true test to evaluate the performance of our model will be to check it on data not used to fit the model
(ii) You can check that we obtained a wide posterior. This is a result of the fact softmax normalizes probability to 1. Therefore, when we used priors on the parameters of 4 species, in reality we can “eliminate” one species” from the problem, in that one of them can be calculated from the other 3 once we know their probabilities (again, they have to sum up to 1!)
(iii) Below is a suggested solution, that does fix the extra parameters to some value, e.g., zero
with pm.Model() as model_sf:
α = pm.Normal('α', mu=0, sigma=2, shape=2)
β = pm.Normal('β', mu=0, sigma=2, shape=(4,2))
α_f = pm.math.concatenate([[0] ,α])
β_f = pm.math.concatenate([np.zeros((4,1)) , β], axis=1)
μ = pm.Deterministic('μ', α_f + pm.math.dot(x_s, β_f))
θ = pm.math.softmax(μ, axis = 1)
yl = pm.Categorical('yl', p=θ, observed=y_s)
idata_sf = pm.sample(2000, target_accept=.92)
data_pred_sf = idata_sf.posterior['μ'].mean(axis=0).mean(axis=0).values
y_pred_sf = [np.exp(point)/np.sum(np.exp(point), axis=0)
for point in data_pred_sf]
res_sf = np.sum(y_s == np.argmax(y_pred_sf, axis=1)) / len(y_s)
print("accuracy is: {:1.3f}".format(res_sf))
accuracy is: 0.973
mu_pred = idata_sf.posterior['μ'].mean(axis=0)
print(np.shape(mu_pred), type(mu_pred))
(2000, 150, 3) <class 'xarray.core.dataarray.DataArray'>
import xarray as xr
def softmax(x):
e_x = np.exp(x)
return e_x.values / np.sum(e_x, axis = 2, keepdims=True).values
res_sm= softmax(mu_pred)
print(np.shape(res_sm))
(2000, 150, 3)
# Average probabilities for prediction
mean_probabilities = np.mean(res_sm, axis=0)
# Uncertainty estimation
lower_bound = np.percentile(res_sm, 2.5, axis=0)
upper_bound = np.percentile(res_sm, 97.5, axis=0)
# class_assignment
class_prediction = np.argmax(y_pred_sf, axis=1)
print(np.shape(class_prediction),np.shape(mean_probabilities),np.shape(lower_bound))
(150,) (150, 3) (150, 3)
print("\n" + "=" * 39)
print("DATA:")
print("=" * 39)
print("class, probabilities, ranges(94%HDI):")
for count, (g, h, i, j, k) in enumerate(zip(y_s, class_prediction, mean_probabilities, lower_bound, upper_bound)):
print(f"data point: {count}, ground-truth: {g}, class: {h}")
for l in range(3):
print(f" idx: {l}, mean prob. {i[l]:.4f}, 94% HDI: [{j[l]:.4f},{k[l]:.4f}]")
if count >= 5:
break
print("=" * 39)
=======================================
DATA:
=======================================
class, probabilities, ranges(94%HDI):
data point: 0, ground-truth: 0, class: 0
idx: 0, mean prob. 0.9905, 94% HDI: [0.9698,0.9991]
idx: 1, mean prob. 0.0095, 94% HDI: [0.0009,0.0302]
idx: 2, mean prob. 0.0000, 94% HDI: [0.0000,0.0000]
data point: 1, ground-truth: 0, class: 0
idx: 0, mean prob. 0.9579, 94% HDI: [0.8851,0.9927]
idx: 1, mean prob. 0.0421, 94% HDI: [0.0073,0.1149]
idx: 2, mean prob. 0.0000, 94% HDI: [0.0000,0.0000]
data point: 2, ground-truth: 0, class: 0
idx: 0, mean prob. 0.9893, 94% HDI: [0.9682,0.9987]
idx: 1, mean prob. 0.0107, 94% HDI: [0.0013,0.0318]
idx: 2, mean prob. 0.0000, 94% HDI: [0.0000,0.0000]
data point: 3, ground-truth: 0, class: 0
idx: 0, mean prob. 0.9845, 94% HDI: [0.9522,0.9981]
idx: 1, mean prob. 0.0155, 94% HDI: [0.0019,0.0478]
idx: 2, mean prob. 0.0000, 94% HDI: [0.0000,0.0000]
data point: 4, ground-truth: 0, class: 0
idx: 0, mean prob. 0.9948, 94% HDI: [0.9820,0.9996]
idx: 1, mean prob. 0.0052, 94% HDI: [0.0004,0.0180]
idx: 2, mean prob. 0.0000, 94% HDI: [0.0000,0.0000]
data point: 5, ground-truth: 0, class: 0
idx: 0, mean prob. 0.9917, 94% HDI: [0.9660,0.9996]
idx: 1, mean prob. 0.0083, 94% HDI: [0.0004,0.0340]
idx: 2, mean prob. 0.0000, 94% HDI: [0.0000,0.0000]
=======================================
Final remarks: Robust Logistic Regression (extra, for the curious…)#
Let’s take the dataset for the species setosa and versicolor only. Let’s complicate the problem by assuming the presence of unusual seros and/or ones in our dataset.
iris = sns.load_dataset("iris")
df = iris.query("species == ('setosa', 'versicolor')")
y_0 = pd.Categorical(df['species']).codes
x_n = 'sepal_length'
x_0 = df[x_n].values
y_0 = np.concatenate((y_0, np.ones(6, dtype=int)))
x_0 = np.concatenate((x_0, [4.2, 4.5, 4.0, 4.3, 4.2, 4.4]))
x_c = x_0 - x_0.mean()
plt.plot(x_c, y_0, 'o', color='k');
plt.xlabel(x_n)
Text(0.5, 0, 'sepal_length')
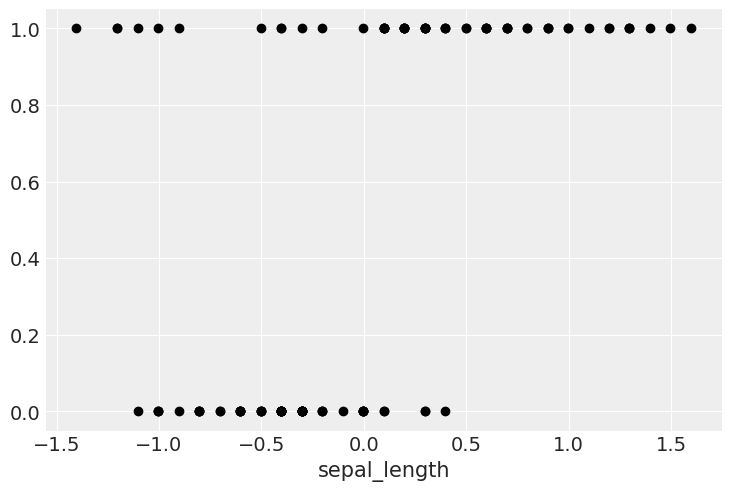
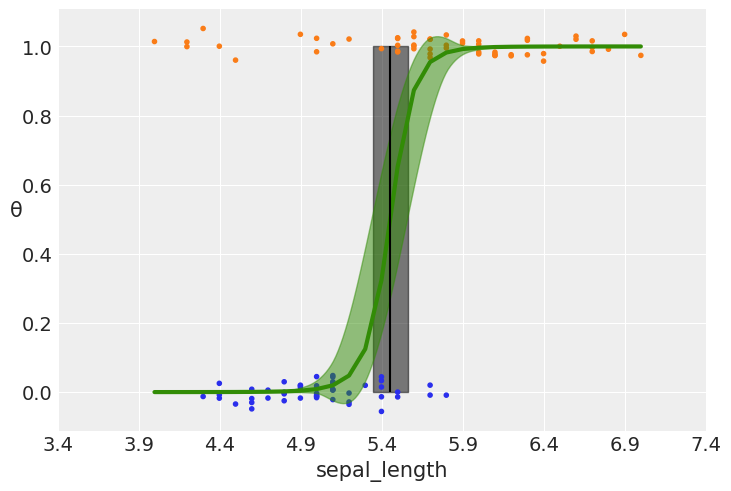
We have some versicolor (category 1) with some unusually short sepal_length…
We can fix this with a mixture model. We say that the outpu variable comes with \(\pi\) probability of random guessing (0.5 chance for category 1 to be indeed 1), and with 1-\(\pi\) probability from a logistic regression model:
\(p= \pi \ 0.5 + (1-\pi) \ logistic(\alpha+X\beta)\)
Notice that when \(\pi=1\), we get \(p=0.5\) (random guess), whereas when \(\pi=0\) we get the logistic regression.
This model can be implemented with a slight modification of what we saw before.
N.B. \(\pi\) is a new variable in our model
with pm.Model() as model_rlg:
α = pm.Normal('α', mu=0, sigma=10)
β = pm.Normal('β', mu=0, sigma=10)
μ = α + x_c * β
θ = pm.Deterministic('θ', pm.math.sigmoid(μ))
bd = pm.Deterministic('bd', -α/β)
π = pm.Beta('π', 1., 1.)
p = π * 0.5 + (1 - π) * θ
yl = pm.Bernoulli('yl', p=p, observed=y_0)
idata_rlg = pm.sample(2000, target_accept=0.95, tune = 2000, return_inferencedata=True)
theta = idata_rlg.posterior['θ'].mean(axis=0).mean(axis=0)
idx = np.argsort(x_c)
np.random.seed(123)
plt.plot(x_c[idx], theta[idx], color='C2', lw=3)
plt.vlines(idata_rlg.posterior['bd'].mean(), 0, 1, color='k')
bd_hpd = az.hdi(idata_rlg.posterior['bd'])
plt.fill_betweenx([0, 1], bd_hpd.bd[0].values, bd_hpd.bd[1].values, color='k', alpha=0.5)
plt.scatter(x_c, np.random.normal(y_0, 0.02),
marker='.', color=[f'C{x}' for x in y_0])
az.plot_hdi(x_c, idata_rlg.posterior['θ'], color='C2') #green band
plt.xlabel(x_n)
plt.ylabel('θ', rotation=0)
# use original scale for xticks
locs, _ = plt.xticks()
plt.xticks(locs, np.round(locs + x_0.mean(), 1))
([<matplotlib.axis.XTick at 0x7d1db3434c10>,
<matplotlib.axis.XTick at 0x7d1db3437be0>,
<matplotlib.axis.XTick at 0x7d1e1727a2c0>,
<matplotlib.axis.XTick at 0x7d1db3d28f40>,
<matplotlib.axis.XTick at 0x7d1dbf50bdf0>,
<matplotlib.axis.XTick at 0x7d1db3b2afe0>,
<matplotlib.axis.XTick at 0x7d1db3d3cac0>,
<matplotlib.axis.XTick at 0x7d1db3b988b0>,
<matplotlib.axis.XTick at 0x7d1db3b993f0>],
[Text(-2.0, 0, '3.4'),
Text(-1.5, 0, '3.9'),
Text(-1.0, 0, '4.4'),
Text(-0.5, 0, '4.9'),
Text(0.0, 0, '5.4'),
Text(0.5, 0, '5.9'),
Text(1.0, 0, '6.4'),
Text(1.5, 0, '6.9'),
Text(2.0, 0, '7.4')])