Bayes Net (or Belief Networks)
Contents
Bayes Net (or Belief Networks)#
Bayesian networks use graphical structures to depict probabilistic relationships between a multitude of variables and facilitate probabilistic inference with these variables. The 1990s witnessed the development of outstanding algorithms for deriving Bayesian networks from observational data.
We focus on intuitive strategies that emphasizes causal learning.
We highlight the connections with the Bayesian approach through straightforward examples. This approach allows us to infer causal relationships from observational data.
Let’s take this problem as our starting point or concrete example to introduce Bayesian Networks through a graph collider.
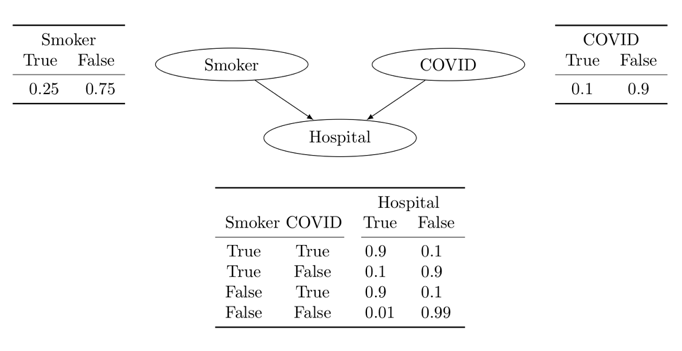
Credits: https://discourse.pymc.io/t/bayes-nets-belief-networks-and-pymc/5150?page=2
Bayes Network Using PyMC#
import pymc as pm
import pytensor.tensor as pt
from pytensor import shared
import numpy as np
lookup_table = shared(np.asarray([
[[.99, .01], [.1, .9]],
[[.9, .1], [.1, .9]]]))
def f(smoker, covid):
return lookup_table[smoker, covid]
with pm.Model() as m_bn:
smoker = pm.Categorical('smoker', [.75, .25])
covid = pm.Categorical('covid', [.9, .1])
hospital = pm.Categorical('hospital', f(smoker, covid))
prior_trace = pm.sample_prior_predictive(100000)
prior_covid = prior_trace.prior['covid'].values
prior_smoker = prior_trace.prior['smoker'].values
prior_hospital = prior_trace.prior['hospital'].values
predict_proba0 = prior_covid[(prior_smoker == 0) & (prior_hospital == 1)].mean()
predict_proba1 = prior_covid[(prior_smoker == 1) & (prior_hospital == 1)].mean()
print(f'P(covid|¬smoking, hospital) is {predict_proba0:1.2f}')
print(f'P(covid|smoking, hospital) is {predict_proba1:1.2f}')
P(covid|¬smoking, hospital) is 0.91
P(covid|smoking, hospital) is 0.49
pgmpy Implementation#
%pip install pgmpy
Requirement already satisfied: pgmpy in /usr/local/lib/python3.10/dist-packages (0.1.25)
Requirement already satisfied: networkx in /usr/local/lib/python3.10/dist-packages (from pgmpy) (3.2.1)
Requirement already satisfied: numpy in /usr/local/lib/python3.10/dist-packages (from pgmpy) (1.25.2)
Requirement already satisfied: scipy in /usr/local/lib/python3.10/dist-packages (from pgmpy) (1.11.4)
Requirement already satisfied: scikit-learn in /usr/local/lib/python3.10/dist-packages (from pgmpy) (1.2.2)
Requirement already satisfied: pandas in /usr/local/lib/python3.10/dist-packages (from pgmpy) (2.0.3)
Requirement already satisfied: pyparsing in /usr/local/lib/python3.10/dist-packages (from pgmpy) (3.1.2)
Requirement already satisfied: torch in /usr/local/lib/python3.10/dist-packages (from pgmpy) (2.2.1+cu121)
Requirement already satisfied: statsmodels in /usr/local/lib/python3.10/dist-packages (from pgmpy) (0.14.1)
Requirement already satisfied: tqdm in /usr/local/lib/python3.10/dist-packages (from pgmpy) (4.66.2)
Requirement already satisfied: joblib in /usr/local/lib/python3.10/dist-packages (from pgmpy) (1.3.2)
Requirement already satisfied: opt-einsum in /usr/local/lib/python3.10/dist-packages (from pgmpy) (3.3.0)
Requirement already satisfied: python-dateutil>=2.8.2 in /usr/local/lib/python3.10/dist-packages (from pandas->pgmpy) (2.8.2)
Requirement already satisfied: pytz>=2020.1 in /usr/local/lib/python3.10/dist-packages (from pandas->pgmpy) (2023.4)
Requirement already satisfied: tzdata>=2022.1 in /usr/local/lib/python3.10/dist-packages (from pandas->pgmpy) (2024.1)
Requirement already satisfied: threadpoolctl>=2.0.0 in /usr/local/lib/python3.10/dist-packages (from scikit-learn->pgmpy) (3.4.0)
Requirement already satisfied: patsy>=0.5.4 in /usr/local/lib/python3.10/dist-packages (from statsmodels->pgmpy) (0.5.6)
Requirement already satisfied: packaging>=21.3 in /usr/local/lib/python3.10/dist-packages (from statsmodels->pgmpy) (24.0)
Requirement already satisfied: filelock in /usr/local/lib/python3.10/dist-packages (from torch->pgmpy) (3.13.3)
Requirement already satisfied: typing-extensions>=4.8.0 in /usr/local/lib/python3.10/dist-packages (from torch->pgmpy) (4.10.0)
Requirement already satisfied: sympy in /usr/local/lib/python3.10/dist-packages (from torch->pgmpy) (1.12)
Requirement already satisfied: jinja2 in /usr/local/lib/python3.10/dist-packages (from torch->pgmpy) (3.1.3)
Requirement already satisfied: fsspec in /usr/local/lib/python3.10/dist-packages (from torch->pgmpy) (2023.6.0)
Requirement already satisfied: nvidia-cuda-nvrtc-cu12==12.1.105 in /usr/local/lib/python3.10/dist-packages (from torch->pgmpy) (12.1.105)
Requirement already satisfied: nvidia-cuda-runtime-cu12==12.1.105 in /usr/local/lib/python3.10/dist-packages (from torch->pgmpy) (12.1.105)
Requirement already satisfied: nvidia-cuda-cupti-cu12==12.1.105 in /usr/local/lib/python3.10/dist-packages (from torch->pgmpy) (12.1.105)
Requirement already satisfied: nvidia-cudnn-cu12==8.9.2.26 in /usr/local/lib/python3.10/dist-packages (from torch->pgmpy) (8.9.2.26)
Requirement already satisfied: nvidia-cublas-cu12==12.1.3.1 in /usr/local/lib/python3.10/dist-packages (from torch->pgmpy) (12.1.3.1)
Requirement already satisfied: nvidia-cufft-cu12==11.0.2.54 in /usr/local/lib/python3.10/dist-packages (from torch->pgmpy) (11.0.2.54)
Requirement already satisfied: nvidia-curand-cu12==10.3.2.106 in /usr/local/lib/python3.10/dist-packages (from torch->pgmpy) (10.3.2.106)
Requirement already satisfied: nvidia-cusolver-cu12==11.4.5.107 in /usr/local/lib/python3.10/dist-packages (from torch->pgmpy) (11.4.5.107)
Requirement already satisfied: nvidia-cusparse-cu12==12.1.0.106 in /usr/local/lib/python3.10/dist-packages (from torch->pgmpy) (12.1.0.106)
Requirement already satisfied: nvidia-nccl-cu12==2.19.3 in /usr/local/lib/python3.10/dist-packages (from torch->pgmpy) (2.19.3)
Requirement already satisfied: nvidia-nvtx-cu12==12.1.105 in /usr/local/lib/python3.10/dist-packages (from torch->pgmpy) (12.1.105)
Requirement already satisfied: triton==2.2.0 in /usr/local/lib/python3.10/dist-packages (from torch->pgmpy) (2.2.0)
Requirement already satisfied: nvidia-nvjitlink-cu12 in /usr/local/lib/python3.10/dist-packages (from nvidia-cusolver-cu12==11.4.5.107->torch->pgmpy) (12.4.127)
Requirement already satisfied: six in /usr/local/lib/python3.10/dist-packages (from patsy>=0.5.4->statsmodels->pgmpy) (1.16.0)
Requirement already satisfied: MarkupSafe>=2.0 in /usr/local/lib/python3.10/dist-packages (from jinja2->torch->pgmpy) (2.1.5)
Requirement already satisfied: mpmath>=0.19 in /usr/local/lib/python3.10/dist-packages (from sympy->torch->pgmpy) (1.3.0)
from pgmpy.models import BayesianNetwork
# Define the structure of the Bayesian Network
model = BayesianNetwork([
('Smoke', 'Hospital'), # Edge from Smoke to Hospital
('Covid', 'Hospital') # Edge from Covid to Hospital
])
# Define the Conditional Probability Distributions (CPDs)
from pgmpy.factors.discrete import TabularCPD
# CPD for Smoke
cpd_smoke = TabularCPD(variable='Smoke', variable_card=2, values=[[0.25], [0.75]], state_names={'Smoke': ['yes', 'no']})
# CPD for Covid
cpd_covid = TabularCPD(variable='Covid', variable_card=2, values=[[0.1], [0.9]], state_names={'Covid': ['yes', 'no']})
# CPD for Hospital
cpd_hospital = TabularCPD(variable='Hospital', variable_card=2,
values=[[0.9, 0.1, 0.9, 0.01], # Probabilities for Hospital=yes
[0.1, 0.9, 0.1, 0.99]], # Probabilities for Hospital=no
evidence=['Smoke', 'Covid'],
evidence_card=[2, 2],
state_names={'Hospital': ['yes', 'no'], 'Smoke': ['yes', 'no'], 'Covid': ['yes', 'no']})
# Add CPDs to the model
model.add_cpds(cpd_smoke, cpd_covid, cpd_hospital)
#-----------------#
#----- NOTES -----#
#-----------------#
# In the CPD for Hospital, the probabilities are ordered according to the Cartesian product of the evidence variables' states,
# i.e., ['Smoke_yes', 'Covid_yes'], ['Smoke_yes', 'Covid_no'], ['Smoke_no', 'Covid_yes'], and ['Smoke_no', 'Covid_no'].
# state_names parameter in each CPD is optional but helps make the output more interpretable.
# This example assumes binary states (yes/no) for simplicity, but pgmpy supports nodes with multiple states.
# The term evidence_card refers to the cardinality of the evidence variables. It specifies the number of states (or distinct values) that each evidence variable can take.
from pgmpy.inference import VariableElimination
# VariableElimination in pgmpy performs exact inference in Bayesian Networks
# by systematically eliminating variables, computing marginal distributions
# of interest or conditional distributions given evidence, optimizing
# computational efficiency through the order of elimination.
inference = VariableElimination(model)
# Probability of Hospitalization given Smoke=yes and Covid=yes
prob_hospital = inference.query(variables=['Hospital'], evidence={'Smoke': 'yes', 'Covid': 'yes'})
print(prob_hospital)
+---------------+-----------------+
| Hospital | phi(Hospital) |
+===============+=================+
| Hospital(yes) | 0.9000 |
+---------------+-----------------+
| Hospital(no) | 0.1000 |
+---------------+-----------------+
# Probability of Covid given Smoke=yes and Hospital=yes
prob_covid = inference.query(variables=['Covid'], evidence={'Smoke': 'yes', 'Hospital': 'yes'})
print(prob_covid)
+------------+--------------+
| Covid | phi(Covid) |
+============+==============+
| Covid(yes) | 0.5000 |
+------------+--------------+
| Covid(no) | 0.5000 |
+------------+--------------+
A more “complex” Network#
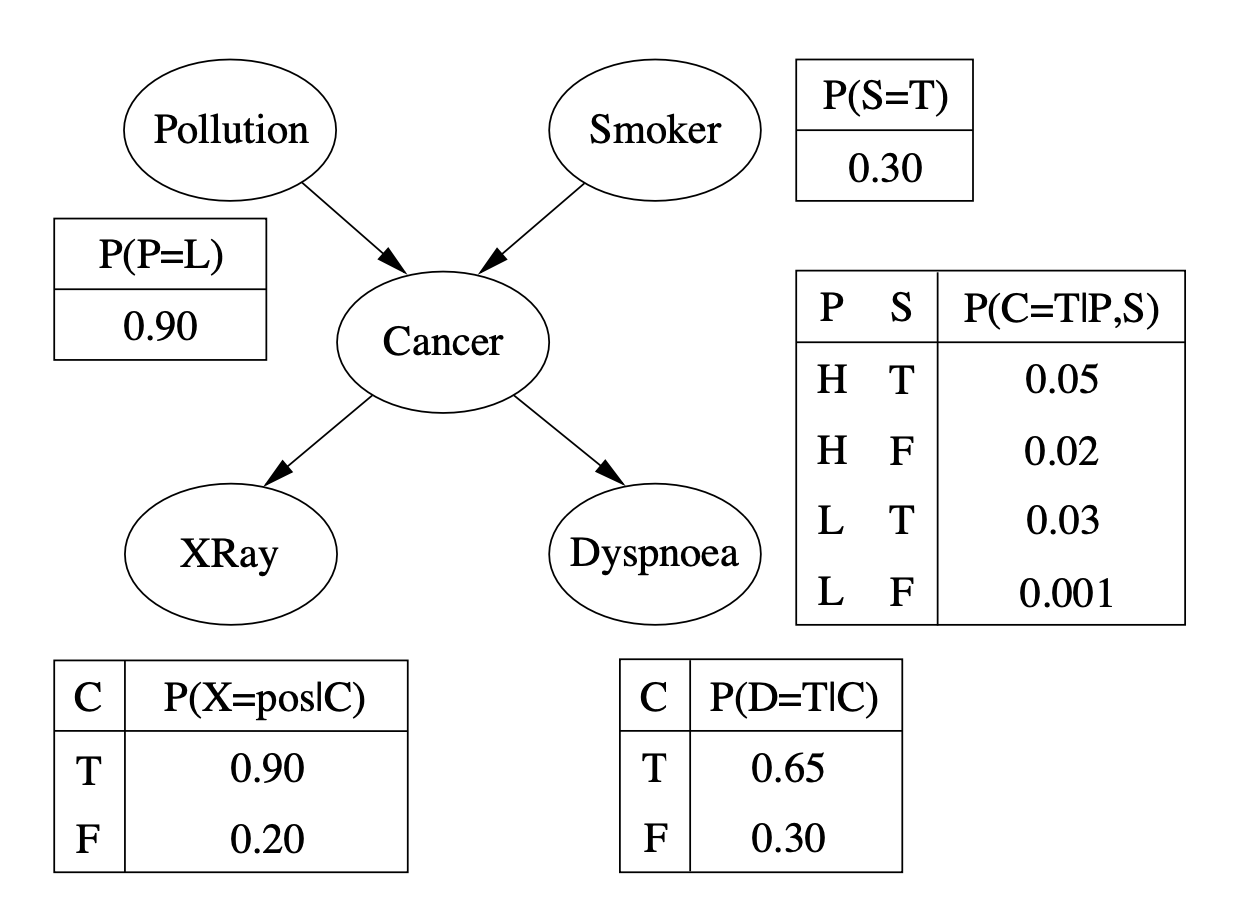
from pgmpy.models import BayesianNetwork
from pgmpy.factors.discrete import TabularCPD
from pgmpy.inference import VariableElimination
# Define the structure of the Bayesian Network
model_complex = BayesianNetwork([('P', 'C'), ('S', 'C'), ('C', 'X'), ('C', 'D')])
# Define the Conditional Probability Distributions (CPDs)
cpd_p = TabularCPD(variable='P', variable_card=2, values=[[0.1], [0.9]], state_names={'P': ['High', 'Low']})
cpd_s = TabularCPD(variable='S', variable_card=2, values=[[0.7], [0.3]], state_names={'S': [False, True]})
cpd_c = TabularCPD(variable='C', variable_card=2,
values=[[0.95, 0.98, 0.97, 0.999],
[0.05, 0.02, 0.03, 0.001]],
evidence=['P', 'S'],
evidence_card=[2, 2],
state_names={'C': [False, True], 'P': ['High', 'Low'], 'S': [False, True]})
cpd_x = TabularCPD(variable='X', variable_card=2,
values=[[0.1, 0.8],
[0.9, 0.2]],
evidence=['C'],
evidence_card=[2],
state_names={'X': ['negative', 'positive'], 'C': [False, True]})
cpd_d = TabularCPD(variable='D', variable_card=2,
values=[[0.35, 0.7],
[0.65, 0.3]],
evidence=['C'],
evidence_card=[2],
state_names={'D': [False, True], 'C': [False, True]})
# Add the CPDs to the model
model_complex.add_cpds(cpd_p, cpd_s, cpd_c, cpd_x, cpd_d)
# Validate the model
assert model_complex.check_model()
# Initialize the inference object
infer = VariableElimination(model_complex)
# Perform query
result = infer.query(variables=['P'], evidence={'D': True, 'S': True})
print(result)
+---------+----------+
| P | phi(P) |
+=========+==========+
| P(High) | 0.0991 |
+---------+----------+
| P(Low) | 0.9009 |
+---------+----------+