Reference: Raschka et al.
https://cfteach.github.io/NNDL_DATA621/referencesmc.html
9. Code examples for Data Preprocessing#
import numpy as np
from IPython.display import Image
%matplotlib inline
9.1. Dealing with missing data#
Identifying missing values in tabular data
import pandas as pd
from io import StringIO
import sys
csv_data = \
'''A,B,C,D
1.0,2.0,3.0,4.0
5.0,6.0,,8.0
10.0,11.0,12.0,'''
# If you are using Python 2.7, you need
# to convert the string to unicode:
if (sys.version_info < (3, 0)):
csv_data = unicode(csv_data)
# StringIO simulates a file=like object in memory,
#like if it was a regular CSV file to read from the hard drive
df = pd.read_csv(StringIO(csv_data))
df
| A | B | C | D | |
|---|---|---|---|---|
| 0 | 1.0 | 2.0 | 3.0 | 4.0 |
| 1 | 5.0 | 6.0 | NaN | 8.0 |
| 2 | 10.0 | 11.0 | 12.0 | NaN |
df.isnull()
| A | B | C | D | |
|---|---|---|---|---|
| 0 | False | False | False | False |
| 1 | False | False | True | False |
| 2 | False | False | False | True |
df.isnull().sum(axis=0)
| 0 | |
|---|---|
| A | 0 |
| B | 0 |
| C | 1 |
| D | 1 |
data = pd.Series([1, np.nan, 3, None, 5])
print(data.isnull())
0 False
1 True
2 False
3 True
4 False
dtype: bool
# access the underlying NumPy array
# via the `values` attribute
df.values
array([[ 1., 2., 3., 4.],
[ 5., 6., nan, 8.],
[10., 11., 12., nan]])
data = np.array([1, np.nan, 3, None], dtype=float)
data
print(np.isnan(data))
[False True False True]
Eliminating training examples or features with missing values
df
| A | B | C | D | |
|---|---|---|---|---|
| 0 | 1.0 | 2.0 | 3.0 | 4.0 |
| 1 | 5.0 | 6.0 | NaN | 8.0 |
| 2 | 10.0 | 11.0 | 12.0 | NaN |
# remove rows that contain missing values
df.dropna(axis=0)
| A | B | C | D | |
|---|---|---|---|---|
| 0 | 1.0 | 2.0 | 3.0 | 4.0 |
# remove columns that contain missing values
df.dropna(axis=1)
| A | B | |
|---|---|---|
| 0 | 1.0 | 2.0 |
| 1 | 5.0 | 6.0 |
| 2 | 10.0 | 11.0 |
# only drop rows where all columns are NaN
df.dropna(how='all')
| A | B | C | D | |
|---|---|---|---|---|
| 0 | 1.0 | 2.0 | 3.0 | 4.0 |
| 1 | 5.0 | 6.0 | NaN | 8.0 |
| 2 | 10.0 | 11.0 | 12.0 | NaN |
# drop rows that have fewer than 4 real values
df.dropna(thresh=4)
| A | B | C | D | |
|---|---|---|---|---|
| 0 | 1.0 | 2.0 | 3.0 | 4.0 |
# only drop rows where NaN appear in specific columns (here: 'C')
df.dropna(subset=['C'])
| A | B | C | D | |
|---|---|---|---|---|
| 0 | 1.0 | 2.0 | 3.0 | 4.0 |
| 2 | 10.0 | 11.0 | 12.0 | NaN |
9.2. Imputing missing values#
# again: our original array
df.values
array([[ 1., 2., 3., 4.],
[ 5., 6., nan, 8.],
[10., 11., 12., nan]])
# impute missing values via the column mean
from sklearn.impute import SimpleImputer
import numpy as np
imr = SimpleImputer(missing_values=np.nan, strategy='mean') #other methods: 'median', 'most_frequent'
imr = imr.fit(df.values)
imputed_data = imr.transform(df.values)
imputed_data
array([[ 1. , 2. , 3. , 4. ],
[ 5. , 6. , 7.5, 8. ],
[10. , 11. , 12. , 6. ]])
df.fillna(df.mean())
| A | B | C | D | |
|---|---|---|---|---|
| 0 | 1.0 | 2.0 | 3.0 | 4.0 |
| 1 | 5.0 | 6.0 | 7.5 | 8.0 |
| 2 | 10.0 | 11.0 | 12.0 | 6.0 |
df
| A | B | C | D | |
|---|---|---|---|---|
| 0 | 1.0 | 2.0 | 3.0 | 4.0 |
| 1 | 5.0 | 6.0 | NaN | 8.0 |
| 2 | 10.0 | 11.0 | 12.0 | NaN |
dg = df.copy()
row, col = (dg == 5.0).stack().idxmax()
print(row, col)
1 A
dg.loc[0,'A'] = None
dg
| A | B | C | D | |
|---|---|---|---|---|
| 0 | NaN | 2.0 | 3.0 | 4.0 |
| 1 | 5.0 | 6.0 | NaN | 8.0 |
| 2 | 10.0 | 11.0 | 12.0 | NaN |
9.3. Handling categorical data#
—– Nominal and ordinal features
import pandas as pd
df = pd.DataFrame([['green', 'M', 10.1, 'class2'],
['red', 'L', 13.5, 'class1'],
['blue', 'XL', 15.3, 'class2']])
df.columns = ['color', 'size', 'price', 'classlabel']
df
| color | size | price | classlabel | |
|---|---|---|---|---|
| 0 | green | M | 10.1 | class2 |
| 1 | red | L | 13.5 | class1 |
| 2 | blue | XL | 15.3 | class2 |
dg = pd.DataFrame([['green','XL','class1'],['yellow','M','class1'],['red','L','class2'],['blue','S','class2']])
dg.columns = ['colors','size','label']
dg
| colors | size | label | |
|---|---|---|---|
| 0 | green | XL | class1 |
| 1 | yellow | M | class1 |
| 2 | red | L | class2 |
| 3 | blue | S | class2 |
mapping = {'XL':4,'L':3,'M':2,'S':1}
dg['size'] = dg['size'].map(mapping)
dg
| colors | size | label | |
|---|---|---|---|
| 0 | green | 4 | class1 |
| 1 | yellow | 2 | class1 |
| 2 | red | 3 | class2 |
| 3 | blue | 1 | class2 |
for k,s in mapping.items():
print(k,s)
XL 4
L 3
M 2
S 1
inverse_mapping = {k:s for s,k in mapping.items()}
inverse_mapping
{4: 'XL', 3: 'L', 2: 'M', 1: 'S'}
dg['size'] = dg['size'].map(inverse_mapping)
dg
| colors | size | label | |
|---|---|---|---|
| 0 | green | XL | class1 |
| 1 | yellow | M | class1 |
| 2 | red | L | class2 |
| 3 | blue | S | class2 |
Mapping ordinal features
size_mapping = {'XL': 3,
'L': 2,
'M': 1}
df['size'] = df['size'].map(size_mapping)
df
| color | size | price | classlabel | |
|---|---|---|---|---|
| 0 | green | 1 | 10.1 | class2 |
| 1 | red | 2 | 13.5 | class1 |
| 2 | blue | 3 | 15.3 | class2 |
inv_size_mapping = {v: k for k, v in size_mapping.items()}
df['size'].map(inv_size_mapping)
| size | |
|---|---|
| 0 | M |
| 1 | L |
| 2 | XL |
9.4. Encoding class labels#
import numpy as np
# create a mapping dict
# to convert class labels from strings to integers
class_mapping = {label: idx for idx, label in enumerate(np.unique(df['classlabel']))}
class_mapping
{'class1': 0, 'class2': 1}
# to convert class labels from strings to integers
df['classlabel'] = df['classlabel'].map(class_mapping)
df
| color | size | price | classlabel | |
|---|---|---|---|---|
| 0 | green | 1 | 10.1 | 1 |
| 1 | red | 2 | 13.5 | 0 |
| 2 | blue | 3 | 15.3 | 1 |
# reverse the class label mapping
inv_class_mapping = {v: k for k, v in class_mapping.items()}
df['classlabel'] = df['classlabel'].map(inv_class_mapping)
df
| color | size | price | classlabel | |
|---|---|---|---|---|
| 0 | green | 1 | 10.1 | class2 |
| 1 | red | 2 | 13.5 | class1 |
| 2 | blue | 3 | 15.3 | class2 |
df['classlabel']
| classlabel | |
|---|---|
| 0 | class2 |
| 1 | class1 |
| 2 | class2 |
df['classlabel'].values
array(['class2', 'class1', 'class2'], dtype=object)
df
| color | size | price | classlabel | |
|---|---|---|---|---|
| 0 | green | 1 | 10.1 | class2 |
| 1 | red | 2 | 13.5 | class1 |
| 2 | blue | 3 | 15.3 | class2 |
from sklearn.preprocessing import LabelEncoder
# Label encoding with sklearn's LabelEncoder
class_le = LabelEncoder()
y = class_le.fit_transform(df['classlabel'].values)
y
array([1, 0, 1])
# reverse mapping
class_le.inverse_transform(y)
array(['class2', 'class1', 'class2'], dtype=object)
9.5. Performing one-hot encoding on nominal features#
df
| color | size | price | classlabel | |
|---|---|---|---|---|
| 0 | green | 1 | 10.1 | class2 |
| 1 | red | 2 | 13.5 | class1 |
| 2 | blue | 3 | 15.3 | class2 |
X = df[['color', 'size', 'price']].values
color_le = LabelEncoder()
X[:, 0] = color_le.fit_transform(X[:, 0])
X
array([[1, 1, 10.1],
[2, 2, 13.5],
[0, 3, 15.3]], dtype=object)
np.shape(X[:, 0])
(3,)
np.shape(X[:, 0].reshape(-1, 1))
(3, 1)
from sklearn.preprocessing import OneHotEncoder
X = df[['color', 'size', 'price']].values
color_ohe = OneHotEncoder()
color_ohe.fit_transform(X[:, 0].reshape(-1, 1)).toarray()
array([[0., 1., 0.],
[0., 0., 1.],
[1., 0., 0.]])
np.shape(color_ohe.fit_transform(X[:, 0].reshape(-1, 1)))
(3, 3)
np.shape(color_ohe.fit_transform(X[:, 0].reshape(-1, 1)).toarray())
(3, 3)
from sklearn.compose import ColumnTransformer
X = df[['color', 'size', 'price']].values
c_transf = ColumnTransformer([ ('onehot', OneHotEncoder(), [0]),
('nothing', 'passthrough', [1, 2])])
c_transf.fit_transform(X).astype(float)
array([[ 0. , 1. , 0. , 1. , 10.1],
[ 0. , 0. , 1. , 2. , 13.5],
[ 1. , 0. , 0. , 3. , 15.3]])
dh = pd.DataFrame([['green',10.2, 1],['blue',8.2, 1],['yellow',9.8, 0]])
dh.columns = ['colors','price','label']
dh
| colors | price | label | |
|---|---|---|---|
| 0 | green | 10.2 | 1 |
| 1 | blue | 8.2 | 1 |
| 2 | yellow | 9.8 | 0 |
dh['colors']
| colors | |
|---|---|
| 0 | green |
| 1 | blue |
| 2 | yellow |
new_var = pd.get_dummies(dh[['colors','price','label']])
df
| color | size | price | classlabel | |
|---|---|---|---|---|
| 0 | green | 1 | 10.1 | class2 |
| 1 | red | 2 | 13.5 | class1 |
| 2 | blue | 3 | 15.3 | class2 |
# one-hot encoding via pandas
pd.get_dummies(df[['price', 'color', 'size']]).astype(int)
| price | size | color_blue | color_green | color_red | |
|---|---|---|---|---|---|
| 0 | 10 | 1 | 0 | 1 | 0 |
| 1 | 13 | 2 | 0 | 0 | 1 |
| 2 | 15 | 3 | 1 | 0 | 0 |
# multicollinearity in get_dummies
pd.get_dummies(df[['price', 'color', 'size']], drop_first=True)
| price | size | color_green | color_red | |
|---|---|---|---|---|
| 0 | 10.1 | 1 | True | False |
| 1 | 13.5 | 2 | False | True |
| 2 | 15.3 | 3 | False | False |
# multicollinearity guard for the OneHotEncoder
color_ohe = OneHotEncoder(categories='auto', drop='first')
c_transf = ColumnTransformer([ ('onehot', color_ohe, [0]),
('nothing', 'passthrough', [1, 2])])
c_transf.fit_transform(X).astype(float)
array([[ 1. , 0. , 1. , 10.1],
[ 0. , 1. , 2. , 13.5],
[ 0. , 0. , 3. , 15.3]])
9.6. Partitioning a dataset into a separate training and test set#
df_wine = pd.read_csv('https://archive.ics.uci.edu/'
'ml/machine-learning-databases/wine/wine.data',
header=None)
df_wine.columns = ['Class label', 'Alcohol', 'Malic acid', 'Ash',
'Alcalinity of ash', 'Magnesium', 'Total phenols',
'Flavanoids', 'Nonflavanoid phenols', 'Proanthocyanins',
'Color intensity', 'Hue', 'OD280/OD315 of diluted wines',
'Proline']
print('Class labels', np.unique(df_wine['Class label']))
df_wine.head()
Class labels [1 2 3]
| Class label | Alcohol | Malic acid | Ash | Alcalinity of ash | Magnesium | Total phenols | Flavanoids | Nonflavanoid phenols | Proanthocyanins | Color intensity | Hue | OD280/OD315 of diluted wines | Proline | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 14.23 | 1.71 | 2.43 | 15.6 | 127 | 2.80 | 3.06 | 0.28 | 2.29 | 5.64 | 1.04 | 3.92 | 1065 |
| 1 | 1 | 13.20 | 1.78 | 2.14 | 11.2 | 100 | 2.65 | 2.76 | 0.26 | 1.28 | 4.38 | 1.05 | 3.40 | 1050 |
| 2 | 1 | 13.16 | 2.36 | 2.67 | 18.6 | 101 | 2.80 | 3.24 | 0.30 | 2.81 | 5.68 | 1.03 | 3.17 | 1185 |
| 3 | 1 | 14.37 | 1.95 | 2.50 | 16.8 | 113 | 3.85 | 3.49 | 0.24 | 2.18 | 7.80 | 0.86 | 3.45 | 1480 |
| 4 | 1 | 13.24 | 2.59 | 2.87 | 21.0 | 118 | 2.80 | 2.69 | 0.39 | 1.82 | 4.32 | 1.04 | 2.93 | 735 |
from sklearn.model_selection import train_test_split
X, y = df_wine.iloc[:, 1:].values, df_wine.iloc[:, 0].values
X_train, X_test, y_train, y_test =\
train_test_split(X, y,
test_size=0.3,
random_state=0,
stratify=y)
9.7. Bringing features onto the same scale#
from sklearn.preprocessing import MinMaxScaler
mms = MinMaxScaler()
X_train_norm = mms.fit_transform(X_train)
X_test_norm = mms.transform(X_test)
from sklearn.preprocessing import StandardScaler
stdsc = StandardScaler()
X_train_std = stdsc.fit_transform(X_train)
X_test_std = stdsc.transform(X_test)
A visual example:
ex = np.array([0, 1, 2, 3, 4, 5])
print('standardized:', (ex - ex.mean()) / ex.std())
# Please note that pandas uses ddof=1 (sample standard deviation)
# by default, whereas NumPy's std method and the StandardScaler
# uses ddof=0 (population standard deviation)
# normalize
print('normalized:', (ex - ex.min()) / (ex.max() - ex.min()))
standardized: [-1.46385011 -0.87831007 -0.29277002 0.29277002 0.87831007 1.46385011]
normalized: [0. 0.2 0.4 0.6 0.8 1. ]
9.8. Selecting meaningful features#
—- Sparse solutions with L1-regularization
For regularized models in scikit-learn that support L1 regularization, we can simply set the penalty parameter to 'l1' to obtain a sparse solution:
from sklearn.linear_model import LogisticRegression
LogisticRegression(penalty='l1')
LogisticRegression(penalty='l1')In a Jupyter environment, please rerun this cell to show the HTML representation or trust the notebook.
On GitHub, the HTML representation is unable to render, please try loading this page with nbviewer.org.
LogisticRegression(penalty='l1')
Applied to the standardized Wine data …
from sklearn.linear_model import LogisticRegression
lr = LogisticRegression(penalty='l1', C=1.0, solver='liblinear', multi_class='ovr')
# Note that C=1.0 is the default. You can increase
# or decrease it to make the regulariztion effect
# stronger or weaker, respectively.
lr.fit(X_train_std, y_train)
print('Training accuracy:', lr.score(X_train_std, y_train))
print('Test accuracy:', lr.score(X_test_std, y_test))
Training accuracy: 1.0
Test accuracy: 1.0
lr.intercept_
array([-1.26352457, -1.21576785, -2.3711671 ])
np.set_printoptions(8)
lr.coef_[lr.coef_!=0].shape
(23,)
lr.coef_
array([[ 1.24574039, 0.18046984, 0.74484363, -1.16251576, 0. ,
0. , 1.16495206, 0. , 0. , 0. ,
0. , 0.55194572, 2.50968308],
[-1.53727681, -0.38718475, -0.99524098, 0.36476702, -0.05956256,
0. , 0.6679675 , 0. , 0. , -1.93383967,
1.23411639, 0. , -2.23183041],
[ 0.13539093, 0.16972595, 0.35779691, 0. , 0. ,
0. , -2.43329298, 0. , 0. , 1.56172158,
-0.81756997, -0.49748177, 0. ]])
np.shape(lr.coef_)
(3, 13)
lr.coef_[1]
array([-1.53727681, -0.38718475, -0.99524098, 0.36476702, -0.05956256,
0. , 0.6679675 , 0. , 0. , -1.93383967,
1.23411639, 0. , -2.23183041])
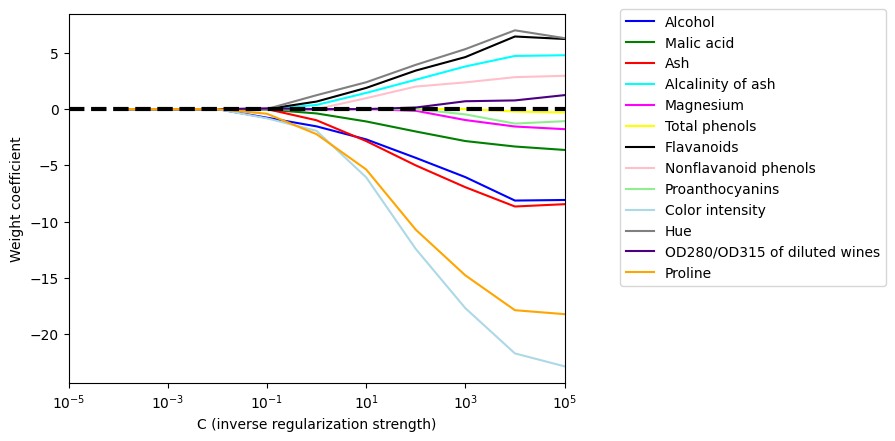
import matplotlib.pyplot as plt
fig = plt.figure()
ax = plt.subplot(111)
colors = ['blue', 'green', 'red', 'cyan',
'magenta', 'yellow', 'black',
'pink', 'lightgreen', 'lightblue',
'gray', 'indigo', 'orange']
weights, params = [], []
for c in np.arange(-4., 6.):
lr = LogisticRegression(penalty='l1', C=10.**c, solver='liblinear',
multi_class='ovr', random_state=0)
lr.fit(X_train_std, y_train)
weights.append(lr.coef_[1])
params.append(10**c)
weights = np.array(weights)
print(np.shape(weights)) #dims of weights (10,13)
# you want to show how each weight (corresponding to a feature) changes vs regularization intensity
for column, color in zip(range(weights.shape[1]), colors):
print(column,color)
plt.plot(params, weights[:, column],
label=df_wine.columns[column + 1],
color=color)
plt.axhline(0, color='black', linestyle='--', linewidth=3)
plt.xlim([10**(-5), 10**5])
plt.ylabel('Weight coefficient')
plt.xlabel('C (inverse regularization strength)')
plt.xscale('log')
plt.legend(loc='upper left')
ax.legend(loc='upper center',
bbox_to_anchor=(1.38, 1.03),
ncol=1, fancybox=True)
#plt.savefig('figures/04_08.png', dpi=300,
# bbox_inches='tight', pad_inches=0.2)
plt.show()
(10, 13)
0 blue
1 green
2 red
3 cyan
4 magenta
5 yellow
6 black
7 pink
8 lightgreen
9 lightblue
10 gray
11 indigo
12 orange
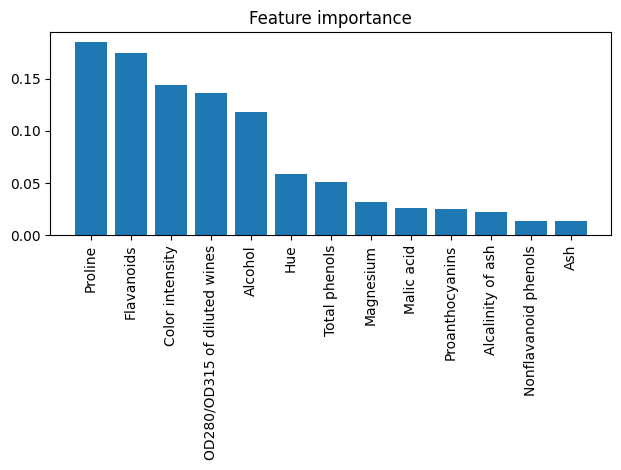
9.9. Assessing feature importance with Random Forests#
from sklearn.ensemble import RandomForestClassifier
feat_labels = df_wine.columns[1:]
forest = RandomForestClassifier(n_estimators=500,
random_state=1)
forest.fit(X_train, y_train)
importances = forest.feature_importances_
indices = np.argsort(importances)[::-1]
for f in range(X_train.shape[1]):
print("%2d) %-*s %f" % (f + 1, 30,
feat_labels[indices[f]],
importances[indices[f]]))
plt.title('Feature importance')
plt.bar(range(X_train.shape[1]),
importances[indices],
align='center')
plt.xticks(range(X_train.shape[1]),
feat_labels[indices], rotation=90)
plt.xlim([-1, X_train.shape[1]])
plt.tight_layout()
# plt.savefig('figures/04_10.png', dpi=300)
plt.show()
1) Proline 0.185453
2) Flavanoids 0.174751
3) Color intensity 0.143920
4) OD280/OD315 of diluted wines 0.136162
5) Alcohol 0.118529
6) Hue 0.058739
7) Total phenols 0.050872
8) Magnesium 0.031357
9) Malic acid 0.025648
10) Proanthocyanins 0.025570
11) Alcalinity of ash 0.022366
12) Nonflavanoid phenols 0.013354
13) Ash 0.013279
from sklearn.feature_selection import SelectFromModel
sfm = SelectFromModel(forest, threshold=0.1, prefit=True)
X_selected = sfm.transform(X_train)
print('Number of features that meet this threshold criterion:',
X_selected.shape[1])
Number of features that meet this threshold criterion: 5
Now, let’s print the 3 features that met the threshold criterion for feature selection that we set earlier (note that this code snippet does not appear in the actual book but was added to this notebook later for illustrative purposes):
for f in range(X_selected.shape[1]):
print("%2d) %-*s %f" % (f + 1, 30,
feat_labels[indices[f]],
importances[indices[f]]))
1) Proline 0.185453
2) Flavanoids 0.174751
3) Color intensity 0.143920
4) OD280/OD315 of diluted wines 0.136162
5) Alcohol 0.118529
—- Example in Python of Feature Importance for Regression problems
from sklearn.ensemble import RandomForestRegressor
from sklearn.datasets import fetch_california_housing
# Load sample data
data = fetch_california_housing()
X, y = data.data, data.target
# Fit Random Forest model
model = RandomForestRegressor(n_estimators=100, random_state=42)
model.fit(X, y)
# Print feature importance
feature_importance = model.feature_importances_
for i, imp in enumerate(feature_importance):
print(f"Feature {data.feature_names[i]}: {imp}")
Feature MedInc: 0.5200367196529164
Feature HouseAge: 0.05296357881747684
Feature AveRooms: 0.04451309296326938
Feature AveBedrms: 0.029298856378707893
Feature Population: 0.03123174949895071
Feature AveOccup: 0.13640641507927073
Feature Latitude: 0.09285575343954347
Feature Longitude: 0.09269383416986465