Reference:
Sebastian Raschka, Yuxi Hayden Liu, and Vahid Mirjalili. Machine Learning with PyTorch and Scikit-Learn: Develop machine learning and deep learning models with Python. Packt Publishing Ltd, 2022.
8. Logistic Regression#
from IPython.display import Image
from IPython.display import display
display(Image(url="https://raw.githubusercontent.com/cfteach/NNDL_DATA621/webpage-src/DATA621/DATA621/images/logreg_flowchart.png", width=700))

Loss function
display(Image(url="https://raw.githubusercontent.com/cfteach/NNDL_DATA621/webpage-src/DATA621/DATA621/images/logreg_loss.png", width=700))
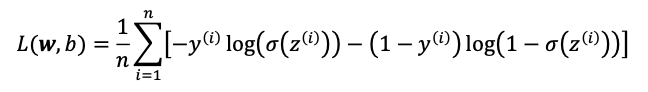
Gradient descent for logistic regression
display(Image(url="https://raw.githubusercontent.com/cfteach/NNDL_DATA621/webpage-src/DATA621/DATA621/images/logreg_gradientdescent.png", width=700))

8.1. Using the Iris data#
import os
import pandas as pd
try:
s = 'https://archive.ics.uci.edu/ml/machine-learning-databases/iris/iris.data'
print('From URL:', s)
df = pd.read_csv(s,
header=None,
encoding='utf-8')
except HTTPError:
s = 'iris.data'
print('From local Iris path:', s)
df = pd.read_csv(s,
header=None,
encoding='utf-8')
df.tail()
From URL: https://archive.ics.uci.edu/ml/machine-learning-databases/iris/iris.data
| 0 | 1 | 2 | 3 | 4 | |
|---|---|---|---|---|---|
| 145 | 6.7 | 3.0 | 5.2 | 2.3 | Iris-virginica |
| 146 | 6.3 | 2.5 | 5.0 | 1.9 | Iris-virginica |
| 147 | 6.5 | 3.0 | 5.2 | 2.0 | Iris-virginica |
| 148 | 6.2 | 3.4 | 5.4 | 2.3 | Iris-virginica |
| 149 | 5.9 | 3.0 | 5.1 | 1.8 | Iris-virginica |
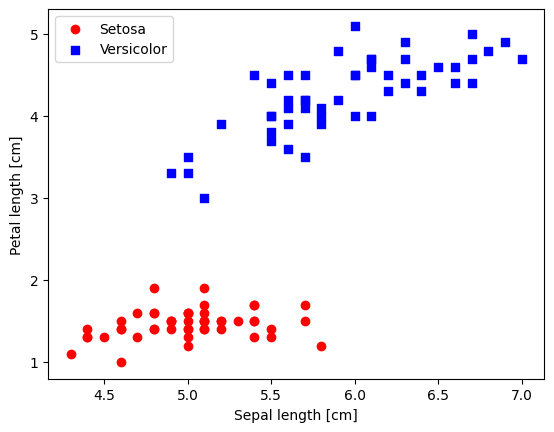
8.2. Plotting the Iris data#
%matplotlib inline
import matplotlib.pyplot as plt
import numpy as np
# select setosa and versicolor
#y = df.iloc[0:100, 4].values
y = df.iloc[:, 4].values
X = df.iloc[:, [0, 2]].values # [0,2] extract sepal length and petal length
# Map y values to 0, 1, or -1
y_mapped = np.select(
[y == 'Iris-setosa', y == 'Iris-versicolor'], # Conditions
[0, 1], # Values to assign if the condition is True
default=-1 # Value to assign if none of the conditions are True
)
mask = (y_mapped == 0) | (y_mapped == 1) # Mask for selecting only 0 and 1 in y_mapped
X_filtered = X[mask]
y_filtered = y_mapped[mask]
# Filter the first 50 occurrences of category 0
mask_0 = (y_filtered == 0)
X_0 = X_filtered[mask_0][:50]
# Filter the first 50 occurrences of category 1
mask_1 = (y_filtered == 1)
X_1 = X_filtered[mask_1][:50]
print(np.shape(X_0))
print(np.shape(X_1))
# plot data
plt.scatter(X_0[:, 0], X_0[:, 1],
color='red', marker='o', label='Setosa')
plt.scatter(X_1[:, 0], X_1[:, 1],
color='blue', marker='s', label='Versicolor')
plt.xlabel('Sepal length [cm]')
plt.ylabel('Petal length [cm]')
plt.legend(loc='upper left')
(50, 2)
(50, 2)
<matplotlib.legend.Legend at 0x7f826489ece0>
8.3. Standardize Data#
This can be very helpful: as we already saw, gradient descent is one of many algorithms that benefit from feature scaling.
X_std = np.copy(X_filtered)
X_std[:,0] = (X_filtered[:,0]-X_filtered[:,0].mean())/X_filtered[:,0].std()
X_std[:,1] = (X_filtered[:,1]-X_filtered[:,1].mean())/X_filtered[:,1].std()
8.4. Logistic Regression#
class LogisticRegressionGD:
"""Gradient descent-based logistic regression classifier.
Parameters
------------
eta : float
Learning rate (between 0.0 and 1.0)
n_iter : int
Passes over the training dataset.
random_state : int
Random number generator seed for random weight
initialization.
Attributes
-----------
w_ : 1d-array
Weights after training.
b_ : Scalar
Bias unit after fitting.
losses_ : list
Log loss function values in each epoch.
"""
def __init__(self, eta=0.01, n_iter=50, random_state=1):
self.eta = eta
self.n_iter = n_iter
self.random_state = random_state
def fit(self, X, y):
""" Fit training data.
Parameters
----------
X : {array-like}, shape = [n_examples, n_features]
Training vectors, where n_examples is the number of examples and
n_features is the number of features.
y : array-like, shape = [n_examples]
Target values.
Returns
-------
self : Instance of LogisticRegressionGD
"""
rgen = np.random.RandomState(self.random_state)
self.w_ = rgen.normal(loc=0.0, scale=0.01, size=X.shape[1])
self.b_ = np.float_(0.)
self.losses_ = []
for i in range(self.n_iter):
net_input = self.net_input(X)
output = self.activation(net_input)
errors = (y - output)
self.w_ += self.eta * X.T.dot(errors) / X.shape[0]
self.b_ += self.eta * errors.mean()
loss = (-y.dot(np.log(output)) - (1 - y).dot(np.log(1 - output))) / X.shape[0]
self.losses_.append(loss.mean())
return self
def net_input(self, X):
"""Calculate net input"""
return np.dot(X, self.w_) + self.b_
def activation(self, z):
"""Compute logistic sigmoid activation"""
return 1. / (1. + np.exp(-np.clip(z, -250, 250)))
def predict(self, X):
"""Return class label after unit step"""
return np.where(self.activation(self.net_input(X)) >= 0.5, 1, 0)
Training the Logistic Regression model
lrgd = LogisticRegressionGD(eta=0.25, n_iter=500, random_state=1)
lrgd.fit(X_std,
y_filtered)
<__main__.LogisticRegressionGD at 0x7f8264853970>
from matplotlib.colors import ListedColormap
def plot_decision_regions(X, y, classifier, resolution=0.02):
# setup marker generator and color map
markers = ('o', 's', '^', 'v', '<')
colors = ('red', 'blue', 'lightgreen', 'gray', 'cyan')
cmap = ListedColormap(colors[:len(np.unique(y))])
# plot the decision surface
x1_min, x1_max = X[:, 0].min() - 1, X[:, 0].max() + 1
x2_min, x2_max = X[:, 1].min() - 1, X[:, 1].max() + 1
xx1, xx2 = np.meshgrid(np.arange(x1_min, x1_max, resolution),
np.arange(x2_min, x2_max, resolution))
lab = classifier.predict(np.array([xx1.ravel(), xx2.ravel()]).T)
lab = lab.reshape(xx1.shape)
plt.contourf(xx1, xx2, lab, alpha=0.3, cmap=cmap)
plt.xlim(xx1.min(), xx1.max())
plt.ylim(xx2.min(), xx2.max())
# plot class examples
for idx, cl in enumerate(np.unique(y)):
plt.scatter(x=X[y == cl, 0],
y=X[y == cl, 1],
alpha=0.8,
c=colors[idx],
marker=markers[idx],
#label=f'Class {cl}',
edgecolor='black')
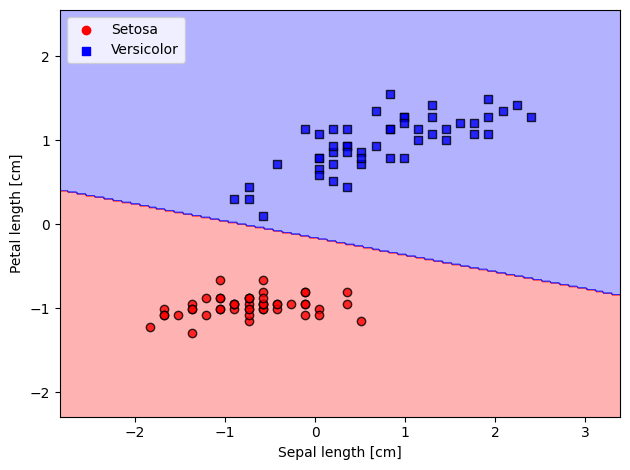
plot_decision_regions(X=X_std,
y=y_filtered,
classifier=lrgd)
# plot data
plt.scatter(X_0[:, 0], X_0[:, 1],
color='red', marker='o', label='Setosa')
plt.scatter(X_1[:, 0], X_1[:, 1],
color='blue', marker='s', label='Versicolor')
plt.xlabel('Sepal length [cm]')
plt.ylabel('Petal length [cm]')
plt.legend(loc='upper left')
plt.tight_layout()
plt.show()
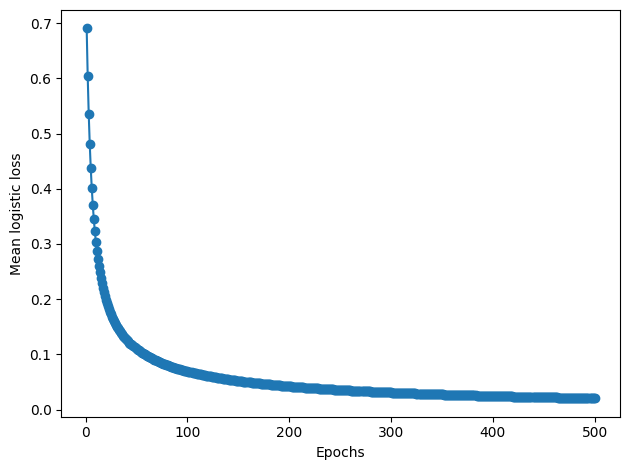
plt.plot(range(1, len(lrgd.losses_) + 1), lrgd.losses_, marker='o')
plt.xlabel('Epochs')
plt.ylabel('Mean logistic loss')
plt.tight_layout()
plt.show()
Accuracy
y_pred = lrgd.predict(X_std)
accuracy = np.sum(y_pred == y_filtered) / len(y_filtered)
print(f"Accuracy: {accuracy * 100:.2f}%")
Accuracy: 100.00%
8.5. Augmenting complexity of decision boundary#
import numpy as np
class LogisticRegressionGD_db:
"""Gradient descent-based logistic regression classifier with polynomial feature augmentation.
Parameters
------------
eta : float
Learning rate (between 0.0 and 1.0)
n_iter : int
Passes over the training dataset.
degree : int, optional (default=2)
Degree of polynomial features (e.g., 2 for quadratic).
random_state : int
Random number generator seed for random weight
initialization.
Attributes
-----------
w_ : 1d-array
Weights after training.
b_ : Scalar
Bias unit after fitting.
losses_ : list
Log loss function values in each epoch.
"""
def __init__(self, eta=0.01, n_iter=50, degree=2, random_state=1):
self.eta = eta
self.n_iter = n_iter
self.degree = degree
self.random_state = random_state
def _augment_features(self, X):
"""Augment feature matrix with polynomial terms up to the specified degree."""
from sklearn.preprocessing import PolynomialFeatures
poly = PolynomialFeatures(self.degree, include_bias=False)
return poly.fit_transform(X)
def fit(self, X, y):
""" Fit training data.
Parameters
----------
X : {array-like}, shape = [n_examples, n_features]
Training vectors, where n_examples is the number of examples and
n_features is the number of features.
y : array-like, shape = [n_examples]
Target values.
Returns
-------
self : Instance of LogisticRegressionGD
"""
# Augment feature space with polynomial features
X_augmented = self._augment_features(X)
rgen = np.random.RandomState(self.random_state)
self.w_ = rgen.normal(loc=0.0, scale=0.01, size=X_augmented.shape[1])
self.b_ = np.float_(0.)
self.losses_ = []
for i in range(self.n_iter):
net_input = self.net_input(X_augmented)
output = self.activation(net_input)
errors = (y - output)
self.w_ += self.eta * X_augmented.T.dot(errors) / X_augmented.shape[0]
self.b_ += self.eta * errors.mean()
loss = (-y.dot(np.log(output)) - (1 - y).dot(np.log(1 - output))) / X_augmented.shape[0]
self.losses_.append(loss.mean())
return self
def net_input(self, X):
"""Calculate net input"""
return np.dot(X, self.w_) + self.b_
def activation(self, z):
"""Compute logistic sigmoid activation"""
return 1. / (1. + np.exp(-np.clip(z, -250, 250)))
def predict(self, X):
"""Return class label after unit step"""
# Augment features before prediction
X_augmented = self._augment_features(X)
return np.where(self.activation(self.net_input(X_augmented)) >= 0.5, 1, 0)
lrgd_db = LogisticRegressionGD_db(eta=0.25, n_iter=500, random_state=1, degree=4)
lrgd_db.fit(X_std, y_filtered)
<__main__.LogisticRegressionGD_db at 0x7f82645748b0>
plot_decision_regions(X=X_std,
y=y_filtered,
classifier=lrgd_db)
# plot data
plt.scatter(X_0[:, 0], X_0[:, 1],
color='red', marker='o', label='Setosa')
plt.scatter(X_1[:, 0], X_1[:, 1],
color='blue', marker='s', label='Versicolor')
plt.xlabel('Sepal length [cm]')
plt.ylabel('Petal length [cm]')
plt.legend(loc='upper left')
plt.tight_layout()
plt.show()
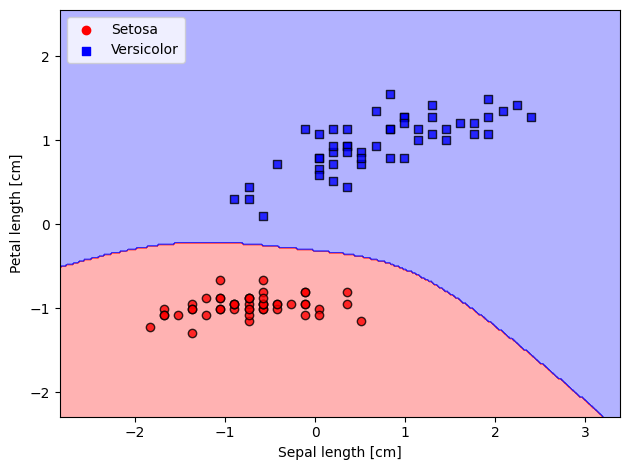
8.6. Adding Regularization#
display(Image(url="https://raw.githubusercontent.com/cfteach/NNDL_DATA621/webpage-src/DATA621/DATA621/images/variance_bias_tradeoff.png", width=700))

class LogisticRegressionGD_reg:
"""Gradient descent-based logistic regression classifier with polynomial feature augmentation and L2 regularization.
Parameters
------------
eta : float
Learning rate (between 0.0 and 1.0)
n_iter : int
Passes over the training dataset.
degree : int, optional (default=2)
Degree of polynomial features (e.g., 2 for quadratic).
alpha : float, optional (default=0.01)
Regularization strength (L2 penalty).
random_state : int
Random number generator seed for random weight
initialization.
Attributes
-----------
w_ : 1d-array
Weights after training.
b_ : Scalar
Bias unit after fitting.
losses_ : list
Log loss function values in each epoch.
"""
def __init__(self, eta=0.01, n_iter=50, degree=2, alpha=0.01, random_state=1):
self.eta = eta
self.n_iter = n_iter
self.degree = degree
self.alpha = alpha # L2 regularization term
self.random_state = random_state
def _augment_features(self, X):
"""Augment feature matrix with polynomial terms up to the specified degree."""
from sklearn.preprocessing import PolynomialFeatures
poly = PolynomialFeatures(self.degree, include_bias=False)
return poly.fit_transform(X)
def fit(self, X, y):
""" Fit training data.
Parameters
----------
X : {array-like}, shape = [n_examples, n_features]
Training vectors, where n_examples is the number of examples and
n_features is the number of features.
y : array-like, shape = [n_examples]
Target values.
Returns
-------
self : Instance of LogisticRegressionGD
"""
# Augment feature space with polynomial features
X_augmented = self._augment_features(X)
rgen = np.random.RandomState(self.random_state)
self.w_ = rgen.normal(loc=0.0, scale=0.01, size=X_augmented.shape[1])
self.b_ = np.float_(0.)
self.losses_ = []
for i in range(self.n_iter):
net_input = self.net_input(X_augmented)
output = self.activation(net_input)
errors = (y - output)
# Gradient descent with L2 regularization for weights
self.w_ += self.eta * (X_augmented.T.dot(errors) / X_augmented.shape[0] - self.alpha * self.w_)
self.b_ += self.eta * errors.mean()
# Compute loss with L2 regularization term
loss = (-y.dot(np.log(output)) - (1 - y).dot(np.log(1 - output))) / X_augmented.shape[0]
loss += (self.alpha / 2.0) * np.sum(self.w_ ** 2) # L2 regularization term
self.losses_.append(loss.mean())
return self
def net_input(self, X):
"""Calculate net input"""
return np.dot(X, self.w_) + self.b_
def activation(self, z):
"""Compute logistic sigmoid activation"""
return 1. / (1. + np.exp(-np.clip(z, -250, 250)))
def predict(self, X):
"""Return class label after unit step"""
# Augment features before prediction
X_augmented = self._augment_features(X)
return np.where(self.activation(self.net_input(X_augmented)) >= 0.5, 1, 0)
lrgd_reg = LogisticRegressionGD_reg(eta=0.25, n_iter=500, random_state=1, degree=4, alpha=0.1)
lrgd_reg.fit(X_std, y_filtered)
<__main__.LogisticRegressionGD_reg at 0x7f8264576bc0>
plot_decision_regions(X=X_std,
y=y_filtered,
classifier=lrgd_reg)
# plot data
plt.scatter(X_0[:, 0], X_0[:, 1],
color='red', marker='o', label='Setosa')
plt.scatter(X_1[:, 0], X_1[:, 1],
color='blue', marker='s', label='Versicolor')
plt.xlabel('Sepal length [cm]')
plt.ylabel('Petal length [cm]')
plt.legend(loc='upper left')
plt.tight_layout()
plt.show()
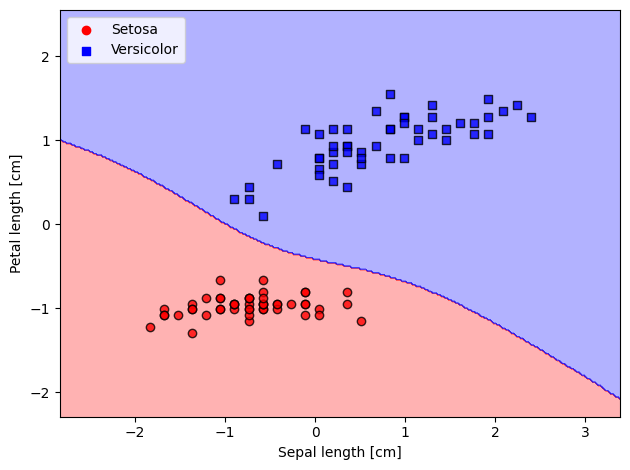
The L2 regularization (used in Ridge regression) tends to make all weights small but non-zero. It is a smooth regularization. Different from L1 (Lasso), which tends to introduce sparsity, i.e., zeroing some weights.
It can be proved that by increasing the regularization term reduce the weights and prevent overfitting. If too high though, can cause bias and underfitting.
8.7. Introduction to Non-Linear Problems#
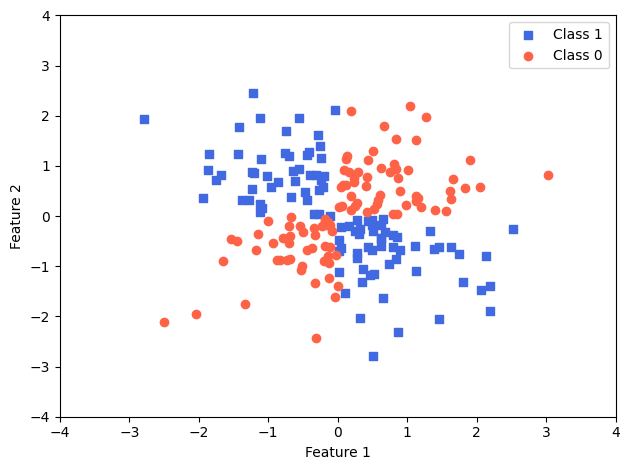
Let’s create another dataset, which is non-linear and corresponding to the logical XOR
np.random.seed(1)
X_xor = np.random.randn(200, 2)
y_xor = np.logical_xor(X_xor[:, 0] > 0,
X_xor[:, 1] > 0)
y_xor = np.where(y_xor, 1, 0)
plt.scatter(X_xor[y_xor == 1, 0],
X_xor[y_xor == 1, 1],
c='royalblue',
marker='s',
label='Class 1')
plt.scatter(X_xor[y_xor == 0, 0],
X_xor[y_xor == 0, 1],
c='tomato',
marker='o',
label='Class 0')
plt.xlim([-4, 4])
plt.ylim([-4, 4])
plt.xlabel('Feature 1')
plt.ylabel('Feature 2')
plt.legend(loc='best')
plt.tight_layout()
plt.show()
lrgd_reg = LogisticRegressionGD_reg(eta=0.25, n_iter=500, random_state=1, degree=4, alpha=0.01)
lrgd_reg.fit(X_xor, y_xor)
<ipython-input-41-cf9e102b21ee>:75: RuntimeWarning: divide by zero encountered in log
loss = (-y.dot(np.log(output)) - (1 - y).dot(np.log(1 - output))) / X_augmented.shape[0]
<__main__.LogisticRegressionGD_reg at 0x7f826474d210>
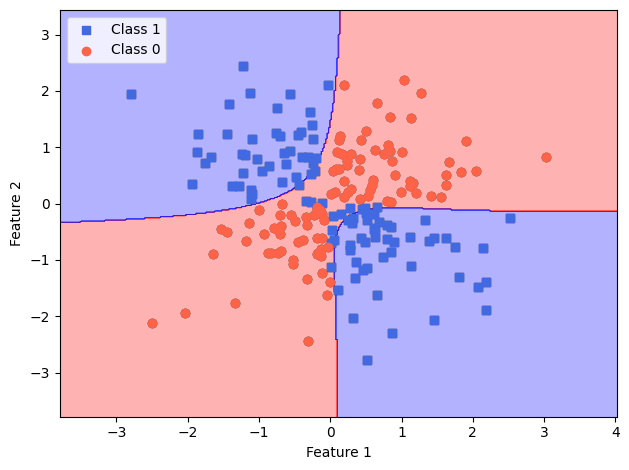
plot_decision_regions(X=X_xor,
y=y_xor,
classifier=lrgd_reg)
# plot data
plt.scatter(X_xor[y_xor == 1, 0],
X_xor[y_xor == 1, 1],
c='royalblue',
marker='s',
label='Class 1')
plt.scatter(X_xor[y_xor == 0, 0],
X_xor[y_xor == 0, 1],
c='tomato',
marker='o',
label='Class 0')
plt.xlabel('Feature 1')
plt.ylabel('Feature 2')
plt.legend(loc='upper left')
plt.tight_layout()
plt.show()