Bayesian A/B Testing
Contents
Bayesian A/B Testing#
!pip install pymc3
Looking in indexes: https://pypi.org/simple, https://us-python.pkg.dev/colab-wheels/public/simple/
Requirement already satisfied: pymc3 in /usr/local/lib/python3.7/dist-packages (3.11.5)
Requirement already satisfied: semver>=2.13.0 in /usr/local/lib/python3.7/dist-packages (from pymc3) (2.13.0)
Requirement already satisfied: scipy<1.8.0,>=1.7.3 in /usr/local/lib/python3.7/dist-packages (from pymc3) (1.7.3)
Requirement already satisfied: dill in /usr/local/lib/python3.7/dist-packages (from pymc3) (0.3.6)
Requirement already satisfied: numpy<1.22.2,>=1.15.0 in /usr/local/lib/python3.7/dist-packages (from pymc3) (1.21.6)
Requirement already satisfied: pandas>=0.24.0 in /usr/local/lib/python3.7/dist-packages (from pymc3) (1.3.5)
Requirement already satisfied: fastprogress>=0.2.0 in /usr/local/lib/python3.7/dist-packages (from pymc3) (1.0.3)
Requirement already satisfied: patsy>=0.5.1 in /usr/local/lib/python3.7/dist-packages (from pymc3) (0.5.3)
Requirement already satisfied: typing-extensions>=3.7.4 in /usr/local/lib/python3.7/dist-packages (from pymc3) (4.1.1)
Requirement already satisfied: theano-pymc==1.1.2 in /usr/local/lib/python3.7/dist-packages (from pymc3) (1.1.2)
Requirement already satisfied: arviz>=0.11.0 in /usr/local/lib/python3.7/dist-packages (from pymc3) (0.12.1)
Requirement already satisfied: deprecat in /usr/local/lib/python3.7/dist-packages (from pymc3) (2.1.1)
Requirement already satisfied: cachetools>=4.2.1 in /usr/local/lib/python3.7/dist-packages (from pymc3) (5.2.0)
Requirement already satisfied: filelock in /usr/local/lib/python3.7/dist-packages (from theano-pymc==1.1.2->pymc3) (3.8.0)
Requirement already satisfied: matplotlib>=3.0 in /usr/local/lib/python3.7/dist-packages (from arviz>=0.11.0->pymc3) (3.2.2)
Requirement already satisfied: setuptools>=38.4 in /usr/local/lib/python3.7/dist-packages (from arviz>=0.11.0->pymc3) (57.4.0)
Requirement already satisfied: xarray-einstats>=0.2 in /usr/local/lib/python3.7/dist-packages (from arviz>=0.11.0->pymc3) (0.2.2)
Requirement already satisfied: xarray>=0.16.1 in /usr/local/lib/python3.7/dist-packages (from arviz>=0.11.0->pymc3) (0.20.2)
Requirement already satisfied: netcdf4 in /usr/local/lib/python3.7/dist-packages (from arviz>=0.11.0->pymc3) (1.6.2)
Requirement already satisfied: packaging in /usr/local/lib/python3.7/dist-packages (from arviz>=0.11.0->pymc3) (21.3)
Requirement already satisfied: cycler>=0.10 in /usr/local/lib/python3.7/dist-packages (from matplotlib>=3.0->arviz>=0.11.0->pymc3) (0.11.0)
Requirement already satisfied: python-dateutil>=2.1 in /usr/local/lib/python3.7/dist-packages (from matplotlib>=3.0->arviz>=0.11.0->pymc3) (2.8.2)
Requirement already satisfied: pyparsing!=2.0.4,!=2.1.2,!=2.1.6,>=2.0.1 in /usr/local/lib/python3.7/dist-packages (from matplotlib>=3.0->arviz>=0.11.0->pymc3) (3.0.9)
Requirement already satisfied: kiwisolver>=1.0.1 in /usr/local/lib/python3.7/dist-packages (from matplotlib>=3.0->arviz>=0.11.0->pymc3) (1.4.4)
Requirement already satisfied: pytz>=2017.3 in /usr/local/lib/python3.7/dist-packages (from pandas>=0.24.0->pymc3) (2022.6)
Requirement already satisfied: six in /usr/local/lib/python3.7/dist-packages (from patsy>=0.5.1->pymc3) (1.15.0)
Requirement already satisfied: importlib-metadata in /usr/local/lib/python3.7/dist-packages (from xarray>=0.16.1->arviz>=0.11.0->pymc3) (4.13.0)
Requirement already satisfied: wrapt<2,>=1.10 in /usr/local/lib/python3.7/dist-packages (from deprecat->pymc3) (1.14.1)
Requirement already satisfied: zipp>=0.5 in /usr/local/lib/python3.7/dist-packages (from importlib-metadata->xarray>=0.16.1->arviz>=0.11.0->pymc3) (3.10.0)
Requirement already satisfied: cftime in /usr/local/lib/python3.7/dist-packages (from netcdf4->arviz>=0.11.0->pymc3) (1.6.2)
Ref.: C. Davidson-Pilon, Bayesian methods for hackers, 2015
Problem: Front-end web developers are interested in which design of their website yields more sales or some other metric of interest create a dataset; they will route some fraction of visitors to site A, and the other fraction to site B, and record if the visit yielded a sale or not. The data is recorded in real-time, and analyzed afterwards.
import pymc3 as pm
from scipy import stats
import numpy as np
import matplotlib.pyplot as plt
# generate our dataset
#these two quantities are unknown to us.
true_p_A = 0.05
true_p_B = 0.04
#notice the unequal sample sizes -- no problem in Bayesian analysis.
N_A = 1500
N_B = 750
#generate some observations
observations_A = stats.bernoulli.rvs(true_p_A, size = N_A)
observations_B = stats.bernoulli.rvs(true_p_B, size = N_B)
print("Obs from Site A: ", observations_A[:30], "...")
print("Obs from Site B: ", observations_B[:30], "...")
Obs from Site A: [0 0 0 0 1 0 0 0 0 0 1 0 0 1 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0] ...
Obs from Site B: [1 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 1 1 0 0 0] ...
Goal: in comparing design A with design B, we want to determine if one of the two is significantly better than the other.
Let’s set up a odel with pymc
# Set up the pymc3 model. Again assume Uniform priors for p_A and p_B.
with pm.Model() as model_all:
p_A = pm.Uniform("p_A", 0, 1)
p_B = pm.Uniform("p_B", 0, 1)
# Define the deterministic delta function. This is our unknown of interest.
delta = pm.Deterministic("delta", p_A - p_B)
# Set of observations, in this case we have two observation datasets.
obs_A = pm.Bernoulli("obs_A", p_A, observed=observations_A)
obs_B = pm.Bernoulli("obs_B", p_B, observed=observations_B)
step = pm.Metropolis() #we will see more next week
trace = pm.sample(20000, step=step)
burned_trace=trace[1000:] #<----- select after burn-in of 1000 samples
/usr/local/lib/python3.7/dist-packages/deprecat/classic.py:215: FutureWarning: In v4.0, pm.sample will return an `arviz.InferenceData` object instead of a `MultiTrace` by default. You can pass return_inferencedata=True or return_inferencedata=False to be safe and silence this warning.
return wrapped_(*args_, **kwargs_)
100.00% [21000/21000 00:17<00:00 Sampling chain 0, 0 divergences]
100.00% [21000/21000 00:08<00:00 Sampling chain 1, 0 divergences]
# compute posteriors on inferred parameters
post_p_A = burned_trace["p_A"]
post_p_B = burned_trace["p_B"]
post_delta = burned_trace["delta"]
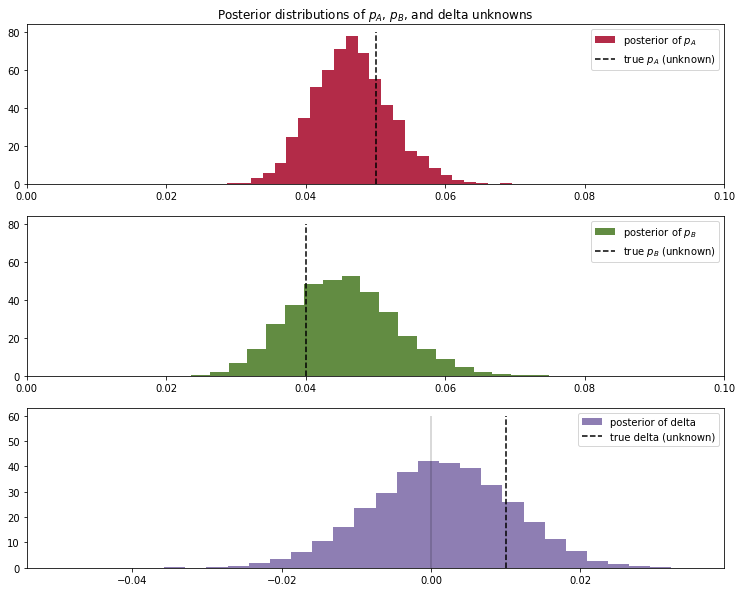
plt.figure(figsize=(12.5, 10))
#histogram of posteriors
ax = plt.subplot(311)
plt.xlim(0, .1)
plt.hist(post_p_A, histtype='stepfilled', bins=25, alpha=0.85,
label="posterior of $p_A$", color="#A60628", density=True)
plt.vlines(true_p_A, 0, 80, linestyle="--", label="true $p_A$ (unknown)")
plt.legend(loc="upper right")
plt.title("Posterior distributions of $p_A$, $p_B$, and delta unknowns")
ax = plt.subplot(312)
plt.xlim(0, .1)
plt.hist(post_p_B, histtype='stepfilled', bins=25, alpha=0.85,
label="posterior of $p_B$", color="#467821", density=True)
plt.vlines(true_p_B, 0, 80, linestyle="--", label="true $p_B$ (unknown)")
plt.legend(loc="upper right")
ax = plt.subplot(313)
plt.hist(post_delta, histtype='stepfilled', bins=30, alpha=0.85,
label="posterior of delta", color="#7A68A6", density=True)
plt.vlines(true_p_A - true_p_B, 0, 60, linestyle="--",
label="true delta (unknown)")
plt.vlines(0, 0, 60, color="black", alpha=0.2)
plt.legend(loc="upper right")
<matplotlib.legend.Legend at 0x7fe98cfef090>
Metrics#
# probability that site A is worse than B
print("probability that site A is worse than B: ", np.mean(post_delta<0))
# probability that site A is better than B
print("probability that site A is better than B: ", np.mean(post_delta>0))
probability that site A is worse than B: 0.44144736842105264
probability that site A is better than B: 0.5585526315789474
#treatment lift
print("treatment lift: ", np.mean((post_p_B-post_p_A)/post_p_A))
treatment lift: -0.01232126175807949
#loss
print("loss: ", np.mean(np.max(post_p_B-post_p_A,0)))
loss: 0.049904685151315624